gff3格式注释文件是最常见的基因注释,(https://archive.broadinstitute.org/annotation/argo/help/gff3.html)
简单来说,gff3是以tab分隔的文本文件,共有9列,对应信息如下:
1、seqname
The name of the sequence. Typically a chromosome or a contig. Argo does not care what you put here. It will superimpose gff features on any sequence you like.
2、source
The program that generated this feature. Argo displays the value of this field in the inspector but does not do anything special with it.
3、feature
The name of this type of feature. The official GFF3 spec states that this should be a term from the SOFA ontology, but Argo does not do anything with this value except display it.
4、start
The starting position of the feature in the sequence. The first base is numbered 1.
5、end
The ending position of the feature (inclusive).
6、score
A score between 0 and 1000. If there is no score value, enter ".".
7、strand
Valid entries include '+', '-', or '.' (for don't know/don't care).
8、frame
If the feature is a coding exon, frame should be a number between 0-2 that represents the reading frame of the first base. If the feature is not a coding exon, the value should be '.'. Argo does not do anything with this field except display its value.
9、GFF3: grouping attributes
Attribute keys and values are separated by '=' signs. Values must be URI encoded.quoted. Attribute pairs are separated by semicolons. Certain, special attributes are used for grouping and identification (See below). This field is the one important difference betweenGFF flavor
(https://archive.broadinstitute.org/annotation/argo/help/gff.html).
在进行生物信息分析的时候,常需要把gene的注释信息(第9列)提取出来附加到差异基因或目的基因的表格结果中,但第9列的注释信息通常较多,且不同基因含部分注释信息不全部一致,一般我们只需要部分重要的a信息,如Dbxref、gene_biotype、deion。
本文以ncbi上发布的人类GRCh38.p7版本注释文件为示例,使用awk命令进行该操作。
(https://www.gnu.org/software/gawk/manual/gawk.html)
1、下载目的物种注释文件:
(ftp://ftp.ncbi.nlm.nih.gov/genomes/all/GCF_000001405.33_GRCh38.p7/GCF_000001405.33_GRCh38.p7_genomic.gff.gz)
然后对 GCF_000001405.33_GRCh38.p7_genomic.gff.gz 进行解压操作,
得到解压文GCF_000001405.33_GRCh38.p7_genomic.gff;
2、查看第9列有哪些注释信息:
$ awk 'BEGIN{FS=OFS="\t"} $3=="gene"{split($9, a, ";"); for(i in a){split(a[i], b, "="); if(++c[b[1]]==1) print b[1]}}' GCF_000001405.33_GRCh38.p7_genomic.gff
运行显示结果有:
ID、Dbxref、Name、deion
gbkey、gene、gene_biotype、pseudo、gene_synonym、partial、start_range、end_range
exception、Note
然后使用以下命令查看gff3文件中的结果:
$ awk -F "\t" '$3=="gene"{print $9}' GCF_000001405.33_GRCh38.p7_genomic.gff | cat -n | less
可以看到
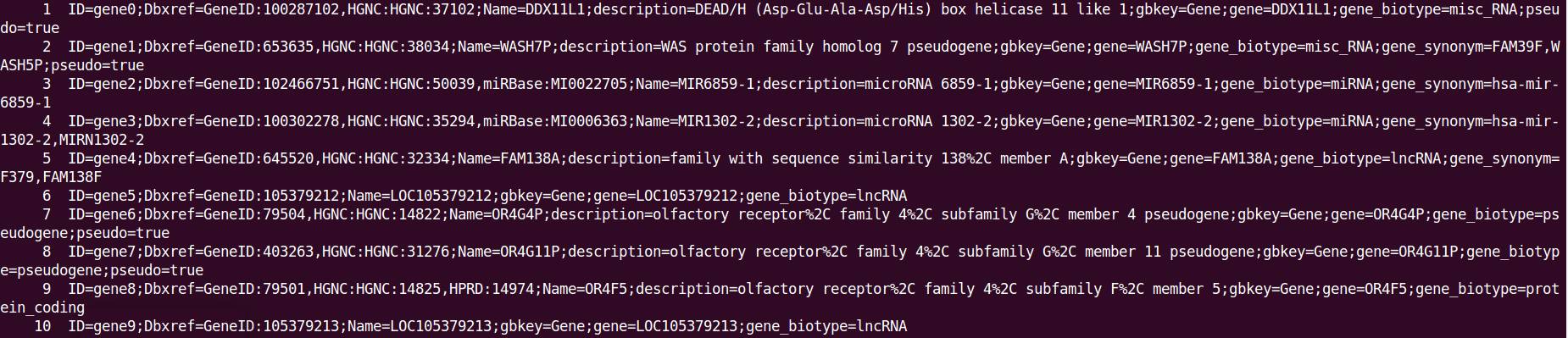
3、下面使用awk进行基因注释信息提取(以提取Dbxref、gene_biotype、deion信息为例):
$ awk 'BEGIN{FS=OFS="\t"} $3=="gene"{print $0}' GCF_000001405.33_GRCh38.p7_genomic.gff |
sed 's/;/\t/g' |
awk 'BEGIN{FS=OFS="\t"} {for(i=1; i<=NF; i++){split($i, a, "=");
b[a[1]]=a[2]}} {print b["Name"],b["Dbxref"],b["gene_biotype"],b["deion"]}
{split("", b, ":")}'
终端显示的提取信息(tab分隔,依次为Name、Dbxref、gene_biotype、deion):
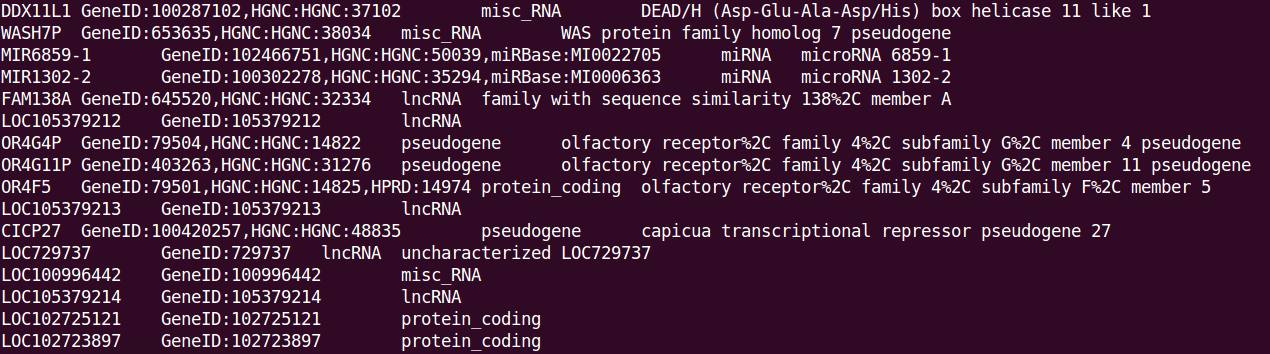
说明:部分基因不包含某些注释信息,如LOC105379212基因没有deion信息,则在对应列为空字符。
4、对应终端打印的提取信息,可以添加表头和生成文件,同时对应部分出现在多个染色体的基因在第1列会重复,请对3中的结果进行以下操作即可:
$ sed ‘1i Name\tDbxref\tgene_biotype\tdeion’ | awk -F “\t” ‘++a[$1]==1’- 本文固定链接: https://maimengkong.com/study/841.html
- 转载请注明: : 萌小白 2022年2月6日 于 卖萌控的博客 发表
- 百度已收录
